-
PDF
- Split View
-
Views
-
Cite
Cite
Catherine I. Dumur, Christine E. Fuller, Tana L. Blevins, Julia C. Schaum, David S. Wilkinson, Carleton T. Garrett, Celeste N. Powers, Clinical Verification of the Performance of the Pathwork Tissue of Origin Test: Utility and Limitations, American Journal of Clinical Pathology, Volume 136, Issue 6, December 2011, Pages 924–933, https://doi.org/10.1309/AJCPDQPFO73SSNFR
Close - Share Icon Share
Abstract
Gene expression–based assays have been introduced into the clinical arena to assist in the diagnosis of poorly differentiated or undifferentiated tumors. The US Food and Drug Administration has cleared the microarray-based Pathwork Tissue of Origin (TOO) Test (Pathwork Diagnostics, Sunnyvale, CA) for the molecular characterization of such challenging specimens. We aimed at verifying the analytic and clinical performance of this test on 43 poorly differentiated and undifferentiated tumor samples, including 6 off-panel cases and 7 cancers of unknown primary (CUP). Our results showed 97% (95% confidence interval, 80.4%–99.8%) agreement between the Pathwork TOO Test result and the complete diagnosis, which included clinical correlations and immunohistochemical staining, after the original diagnosis. We concluded that for off-panel and CUP samples, the tissue type and the cell type may be confounded by the Pathwork TOO Test and that careful clinicopathologic assessment is needed when interpreting results from this helpful ancillary tool for pathologists.
Poorly differentiated or undifferentiated tumors may manifest as primary or metastatic lesions and often prove formidable diagnostic dilemmas from a histopathologic perspective. On rare occasions, the accurate identification of the primary tumor cannot be ascertained, leading to a diagnosis of cancer of unknown origin, which is associated with a poor prognosis.1,2 Pathologists have routinely used immunohistochemical algorithms to assess the tissue type present in such specimens; based on phenotypic characteristics revealed by panels of lineage- and cell-specific antibodies,3 an accurate diagnosis can be made in 75% of these cases.4 Unfortunately, the remaining 25% are not classified, leaving pathologists and clinicians frustrated about the additional patient workup or treatment modalities to pursue. Recently, microarray-based gene expression profiling has shown promise in providing a molecular means of tumor classification.5–7 Platforms using support vector machine classifiers have been demonstrated to be of potential value in classifying cancers of unknown primary (CUP), revealing 89% accuracy when classifying 13 different tissue types.8
Newly developed molecular assays have been introduced into the clinical arena to assist in the diagnosis of difficult-to-characterize tumors and CUP specimens. Such methods include the measurement of transcripts isolated from fresh frozen or formalin-fixed, paraffin-embedded specimens by quantitative real-time polymerase chain reaction,9–11 by the use of microarray platforms,12–15 or by the characterization of microRNA profiles.16,17 Of all these commercially available assays, the Pathwork Tissue of Origin (TOO) Test (Pathwork Diagnostics, Sunnyvale, CA) is the only one that has been cleared by the US Food and Drug Administration (FDA) as an in vitro diagnostic multivariate index assay.
The Pathwork TOO Test is a microarray-based gene expression assay intended to determine the similarity of a tumor specimen to 15 known tissue types of high incidence in metastatic solid tumors. The assay measures the expression of 1,550 genes in each sample, then standardized expression data are sorted through a classification algorithm by comparing expression profiles with the 15 tissue types covered by the test. For such comparisons, 15 similarity scores (ranging from 0 to 100 per class and adding up to 100 for all 15 classes) are generated, each reflecting a similarity to 1 of the 15 tissue types covered by the assay. The analytic validation of the test using frozen tissue samples, including the reproducibility of the assay across 4 different laboratories,18 and the clinical validation based on 547 specimens12 have been previously reported. The assay has been shown to perform with 87.8% sensitivity and 99.4% specificity compared with the reference diagnosis.
According to the Clinical Laboratory Improvement Amendments of 1988 and as recommended by the Clinical and Laboratory Standards Institute,19 the analytic and clinical performance of FDA-cleared assays must be confirmed before implementation of the test. In the present study, we examined the ability of the Pathwork TOO Test to identify the primary tissue type in samples using the largest series published thus far of poorly differentiated and undifferentiated primary and metastatic tumors.
Materials and Methods
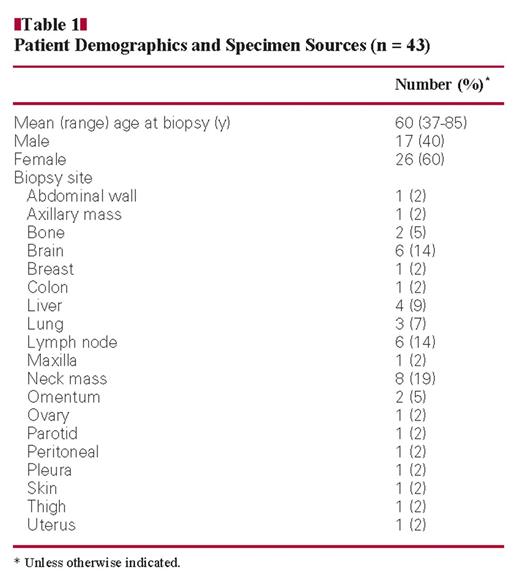
Tissue Specimens
Residual tissue samples from diagnosis, along with patient informed consent, were obtained and banked under a Virginia Commonwealth University (Richmond) Institutional Review Board–approved protocol. Tissue from each surgical pathology specimen was snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. A total of 43 tumor specimens were identified on the basis that their original histopathologic diagnosis met the following criteria: poorly to moderately differentiated primary tumors, including, but not limited to, 15 tissue types covered by the Pathwork TOO Test and poorly to moderately differentiated metastatic tumors, with 1 or 2 possible primary sites, including metastases from an unknown origin. The clinical features and patient demographics are listed in Table 1. Histopathologic scoring of standard features (tumor content, stroma contribution, and presence of necrosis) was performed by a pathologist (C.T.G.) on H&E-stained frozen sections adjacent to, above, and below the tissue used for RNA isolation.
Clinical Verification Study Design
The goal of this study was to assess the clinical performance of the FDA-cleared Pathwork TOO Test in selected cases from our institution that were deemed difficult to diagnose. To further assess the performance of this test, we decided to include 6 (14%) specimens with diagnoses not covered by the test panel of 15 tissue types (“off-panel” cases) and 7 (16%) specimens diagnosed as CUP. The remaining 30 (70%) specimens corresponded to 20 (67%) metastases and 10 (33%) primary tumors, with at least 1 of the possible diagnoses corresponding to 1 tissue type covered by the test panel.
Total RNA Isolation
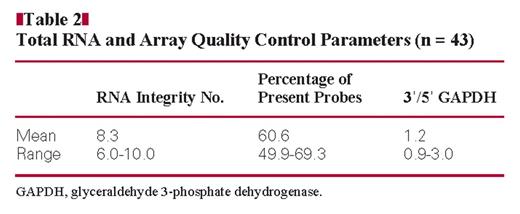
Total RNA was extracted and the quality evaluated using a sample processing method previously established in our laboratory.20 Total RNA purity was judged by spectrophotometry at 260, 270, and 280 nm. RNA integrity and complementary DNA and complementary RNA synthesis products were assessed by running 1 μL of every sample in RNA 6000 Nano LabChips on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). RNA samples showing RNA integrity number values of 6.0 or more were included in the study18Table 2.
Gene Expression Microarray Analysis
The total RNA samples were analyzed on Pathchip arrays following the standardized Affymetrix (Santa Clara, CA) protocol, as described elsewhere.18 Briefly, 1 μg of total RNA was transcribed and further labeled using the GeneChip One-Cycle cDNA (complementary DNA) Synthesis Kit (manufactured by Invitrogen, San Diego, CA, for Affymetrix) and the GeneChip IVT Labeling Kit (Affymetrix). Fifteen micrograms of the fragmented labeled complementary RNA was hybridized to Pathchip microarrays. After scanning the chips on the Affymetrix GeneChip Scanner 3000Dx, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats). The overall quality of each array was assessed by monitoring the 3′/5′ ratios for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the percentage of “present” genes (%P), and by the TOO Test data verification algorithm. Arrays showing GAPDH 3′/5′ ratios of 3.0 or more, %P of 40% or more,21 and “acceptable” data quality on the Pathwork TOO Test report were included in the study (Table 2).
Pathwork TOO Test
The Pathwork TOO Test is an in vitro diagnostic test cleared by the FDA for evaluating the TOO in poorly differentiated or undifferentiated tumors. The analytic performance and the clinical validation of this test have been described.12,18 Briefly, this microarray-based gene expression test quantifies the similarity of tumor specimens to 15 known primary locations, including bladder, breast, colorectal, gastric, germ cell, hepatocellular, kidney, non–small cell lung, non-Hodgkin lymphoma, melanoma, ovarian, pancreatic, prostate, soft tissue sarcoma, and thyroid, and excluding cholangiocarcinoma, uterus, head and neck squamous cell carcinoma (HNSCC), etc. Gene expression data from each .CEL file is further normalized and used to perform pairwise comparisons for each pair of 15 tissues on the test panel, as previously described.18 The results are reported as 15 similarity scores (SS), 1 for each tissue included in the test panel. The SS are probability-based, with a reported range from 0 to 100, and the sum of all 15 possible SS per specimen equals 100. An SS of 30 or more indicates the presence of a given tissue in the specimen, and an SS of 5 or less indicates the absence of a given tissue type in the specimen. SS between 5 and 30 are considered indeterminate.18 These criteria were used to make a tissue determination for each specimen analyzed in the present study.
Immunohistochemical Studies and Clinical History Review
Discordant results between the Pathwork TOO Test and the original diagnosis were further evaluated by immunohistochemical analysis with the following antibodies: CDX2, carcinoembryonic antigen, HMB-45, MART-1, thyroid transcription factor (TTF)-1, and tyrosinase, in addition to those previously used during the original diagnostic workup. In addition, clinical histories were reviewed and computed tomography (CT) scan images were obtained and correlated, when available.
Statistical Analyses
Agreement confidence intervals were calculated using the lower and upper limits of the 95% confidence interval (CI) for proportions, according to the Wilson procedure22 with a correction for continuity.23
Results
RNA Sample and Microarray Quality
All 43 samples passed the total RNA and array quality control criteria described in the “Materials and Methods” section (Table 2) and were deemed acceptable by the Pathwork TOO Test data verification algorithm. However, only 40 contained more than 60% tumor and less than 20% necrosis, as recommended by the Pathwork TOO Test specifications. Therefore the 3 cases not meeting this specification were excluded from consideration regarding the accuracy of the TOO Test, although the results from their study were examined with regard to the impact of low percentage of tumor as a potential confounding factor (see “Performance of the Pathwork TOO Test on Off-Panel and Inadequate Samples”).
Performance Characteristics of the Pathwork TOO Test
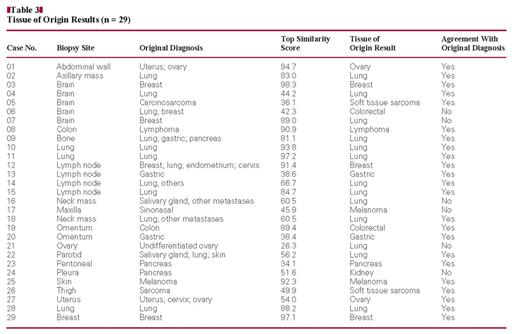
Of the 40 samples included in this study, 35 had a diagnosis other than CUP. Of these 35 specimens, 29 were diagnosed with at least 1 TOO included in the 15-tissue panel covered by the test. The Pathwork TOO Test results and the agreement with the original diagnosis are shown in Table 3. Thus, the 29 tumors included a total of 10 different tissue types of the 15 possible types covered by the test. There was agreement between the Pathwork TOO Test and the initial histopathologic diagnosis in 79% of the cases (23/29; 95% CI, 59.7%–91.3%).
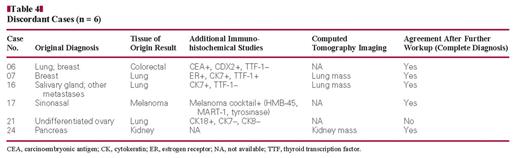
The 6 discordant cases (relative to the original diagnosis) were further investigated by performing immunohistochemical analysis and analyzing the clinical history obtained after the initial histopathologic diagnosis, including imaging results, to compare the Pathwork TOO Test results with the “final diagnosis” made with this additional workup Table 4. One of the discordant cases (case 21) corresponded to an adnexal mass diagnosed as an undifferentiated carcinoma, not further classifiable. The specimen showed an immunohistochemical staining pattern supporting the interpretation of an epithelial neoplasm primary in the ovary. The Pathwor k TOO Test result for this case was “indeterminate,” although its top SS score (which was <30) favored a “lung” origin. For purposes of this study, however, we considered that this case represented a misclassification by the Pathwork TOO Test. However, in the remaining 5 discordant cases, we found that results from subsequent clinical and immunohistochemical evaluation resulted in histopathologic diagnosis that concurred with Pathwork TOO Test results.
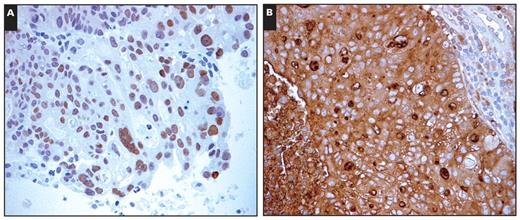
Case 06 manifested with a right parietal brain tumor with suspicion of lung lesions. The brain mass was originally reported as metastatic adenocarcinoma, favoring a lung origin. The Pathwork TOO Test indicated a “colorectal” origin with a top SS of more than 30, and further immunohistochemical studies showed positive staining with carcinoembryonic antigen and CDX2 Image 1, suggesting intestinal differentiation.
(Case 06) Sections of a selected formalin-fixed, paraffin-embedded block from right parietal brain tumor. Immunohistochemical studies revealed positive nuclear staining for CDX2 (A, ×400) and positive cytoplasmic staining for carcinoembryonic antigen (B, ×400).
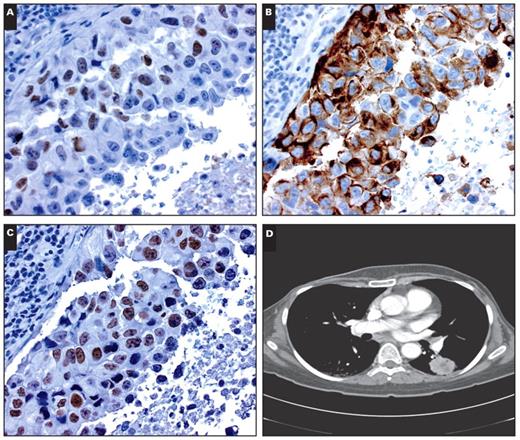
Case 07 corresponded to a solitary enhancing right parieto-occipital lesion manifesting 4 years after mastectomy in a 47-year-old woman with a history of breast carcinoma, liposarcoma, and nerve sheath tumor. A preliminary diagnosis of poorly differentiated metastatic adenocarcinoma suggesting a breast origin was made based on an immunostaining profile showing positivity for estrogen receptor Image 2A and cytokeratin (CK) 7 Image 2B. Surprisingly, the Pathwork TOO Test indicated a lung origin with a top SS near 90. While unusual, primary lung cancer cases showing estrogen receptor positivity have been reported.24 Additional radioimaging studies revealed a large lung mass and scattered small nodular opacities Image 2D, and subsequent immunohistochemical assessment of TTF-1 was likewise positive Image 2C, indicative of a lung primary. Subsequently, the patient underwent surgery, with stabilization of the lung tumor, and no additional new lesions were found during her current chemotherapeutic regimen of carboplatin and paclitaxel.
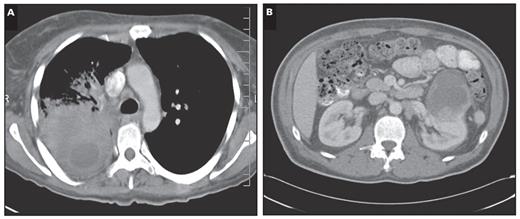
Case 16 corresponded to a submandibular mass originally diagnosed as an undifferentiated carcinoma. Furthermore, the original pathology report stated that this tumor was best classified as an undifferentiated, large cell–type tumor of the salivary gland, although a metastasis could not be excluded. The Pathwork TOO Test result indicated a lung origin (top SS, 60.5). Five months later, a dominant heterogeneous necrotizing tumor-consolidative process replacing the vast majority of the right upper lobe Image 3A was found by CT scan.
Case 24 corresponded to a pleural lesion in a 46-year-old man with pulmonary nodules and in whom suspicion of a pancreatic mass was raised by radiologic imaging. The pathologist was asked to exclude lung carcinoma. The specimen showed an immunohistochemical staining pattern that excluded lung and prostate primary sites and was subsequently diagnosed as a moderate to poorly differentiated metastatic adenocarcinoma, favoring pancreatic primary. The Pathwork TOO Test resulted in a “kidney” type with a top SS of 51.6. A 7-cm3 mass originating from the anterior upper pole of the left kidney with mixed attenuation with perinephric stranding and a small amount of fluid was identified 1 month later by CT Image 3B. Moreover, although the renal mass displaced the tail of the pancreas superiorly, it did not appear to originate from the pancreas as a fat plane was clearly detectable between the pancreas and the renal tumor.
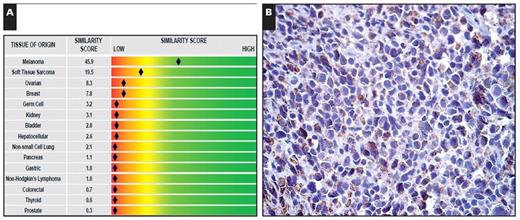
Last, case 17 corresponded to a tumor mass from the right maxilla originally diagnosed as an undifferentiated sinonasal carcinoma with widespread necrosis. The Pathwork TOO Test indicated a “melanoma” type with a top SS of 45.9 Image 4A, and further immunohistochemical studies showed positive staining with a melanoma cocktail containing HMB-45, MART-1 (melan-A), and tyrosinase Image 4B. In this case, the Pathwork TOO Test was identifying a particular cell (melanocytic), rather than a TOO (such as neural crest or skin).
Taking into account the information from subsequent clinical history and immunohistochemical studies in these cases, we found 97% agreement (28/29; 95% CI, 80.4%–99.8%) between the Pathwork TOO Test result and the final diagnosis.
Evaluation of CUP Tumors
Among the 40 samples included in this study, 5 were originally diagnosed as CUP. Although a direct correlation between the Pathwork TOO Test result and the diagnosis could not be assessed, a clinicopathologic correlation was performed to evaluate the usefulness of the TOO Test in these difficult cases. These 5 specimens were obtained from the following biopsy sites: 1 brain lesion, 2 neck masses, 1 cervical lymph node, and 1 liver mass. All 5 top SS were greater than 30, corresponding to 4 lung and 1 “liver” result, respectively. The brain lesion had the morphologic appearance and immunohistochemical profile most consistent with a poorly differentiated squamous cell carcinoma, and a chest CT showed a dominant mass within the right lower lobe that abutted the right hilum with noted nodularity along the right oblique fissure and multiple additional nodules throughout both lungs, consistent with malignancy.
(Case 07) Sections of a selected formalin-fixed, paraffin-embedded block from a right parieto-occipital lesion. Immunohistochemical studies revealed positive nuclear staining for estrogen receptor (A, ×400), positive cytoplasmic staining for cytokeratin 7 (B, ×400), and positive nuclear staining for thyroid transcription factor-1 (C, ×400). D, A chest computed tomography scan revealed a 3-cm mass with an irregular border and central heterogeneous density suggestive of necrosis within the superior segment of the left lower lobe of the lung.
The 2 neck masses had the histologic appearance of squamous cell carcinoma, as did the cervical lymph node metastasis, and they were further classified by the pathologist as HNSCC, with an increased tracer uptake identified by positron emission tomography in a focal region of the left medial palatine tonsil, suggesting this was the primary for the cervical lymph node metastasis. Because HNSCC is not a tissue type covered by the Pathwork TOO Test, a lung result may suggest that the assay is identifying a histologic squamous cell type, often seen in non–small cell lung carcinomas.
The liver mass consisted of a poorly differentiated adenocarcinoma with an immunoprofile showing strong positivity for epithelial membrane antigen, Cam 5.2, and MOC-31 and negativity for AE1/AE3, CK7, CK19, CK20, Hep Par-1, and TTF-1. The liver result yielded by the Pathwork TOO Test on this case may suggest that the assay is identifying the biopsy site rather than the tumor or that the actual tumor type is not covered by the 15-tissue panel, such as a possible cholangiocarcinoma primary.
Computed tomography scan results. A (Case 16), A dominant heterogeneous necrotizing tumor-consolidative process in the right upper lobe of the lung. B (Case 24), A 7-cm3 mass on the left kidney.
(Case 17) Melanoma cell type identification. A “melanoma” expression profile according to the Pathwork Tissue of Origin Test report (A) and positive immunohistochemical staining using a melanoma cocktail containing HMB-45, MART-1 (melan-A), and tyrosinase antibodies (B, ×400).
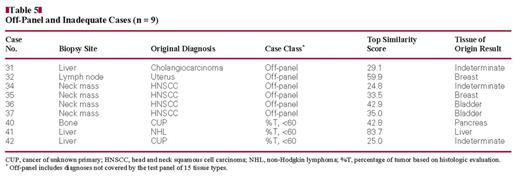
Performance of the Pathwork TOO Test on Off-Panel and Inadequate Samples
When using the Pathwork TOO Test, one important caveat is that the actual TOO for a given tumor specimen might not be covered by the 15 tissue types included in the test panel. To assess the performance of this test on such cases, we tested 6 tumor specimens diagnosed with a TOO type not covered by the assay (off-panel cases). The Pathwork TOO Test results for these cases are shown in Table 5. Of 6 cases, 2 were classified as indeterminate, which would be considered the expected result for off-panel cases. However, 4 of the 6 cases gave TOO Test results indicating TOOs that did not match the clinical and histopathologic findings for these off-panel tumors. Four of the cases were HNSCC. It is interesting that 2 of the 4 gave a result of “bladder” TOO results. However, even this information proved helpful in case 37, in which a possible thyroid cancer was expected on a neck mass biopsy based on the clinical history of the patient. Thus, the result from the TOO Test was helpful in excluding thyroid (SS = 1.3).
When assessing biopsy material, there is a possibility of finding that the specimen includes less than 60% of tumor content. That was the case for 3 of the 43 cases included in this study. Of the 3 inadequate samples corresponding to a CUP specimen, 1 (case 42) had a result of a top SS less than 30, corresponding to an indeterminate TOO result; while a liver biopsy (case 41) yielded a very high SS (83.7) corresponding to liver, indicating that the biopsy site itself was being identified, rather than the actual tumor type, owing to the poor representation of tumor cells in the sample (Table 5).
Discussion
The microarray-based Pathwork TOO Test has been analytically18 and clinically12 validated, resulting in clearance from the FDA as an in vitro diagnostic multivariate index assay. A few previous studies have reported the usefulness of the microarray-based Pathwork TOO Test in assisting the diagnosis of a series of 15 brain metastases with known primaries25 and a series of 21 CUPs.26
Our study constitutes the clinical verification of this FDA-cleared test, using the largest reported series of poorly differentiated and undifferentiated primary and metastatic tumor specimens. Our results showed a 79% agreement between the Pathwork TOO Test result and the original diagnosis for 29 adequate samples with possible diagnoses among the 15 tissue types covered by the assay of a total of 43 samples analyzed. However, we found 97% agreement when account was taken of subsequent clinical (including imaging results) and immunohistochemical findings for the 6 discordant cases. These results show higher performance of the Pathwork TOO Test compared with well-established immunohistochemical algorithms, the latter ranging in reported accuracy from 75% to 88%.4,27,28 For adequate specimens, the performance of the Pathwork TOO Test was verified and was congruent with the reported clinical validation.12
More important, the results of the Pathwork TOO Test were in agreement with imaging and immunohistochemical results obtained several months after the original diagnosis. The imaging-based tests were performed in an attempt to assess the TOO of the tumor because the original diagnosis was not decisive. Taking this timeline into account, the usefulness of the Pathwork TOO Test is underscored because the results of this study show that a TOO call could have been obtained several months in advance, preventing the need for further immunohistochemical and imaging testing and increasing the chances of therapy success in patients given an accurate diagnosis with the correct cancer type.1,11 In addition, prompt and accurate diagnosis of cancer type would result in a reduction of the total cost of diagnosing poorly differentiated or undifferentiated CUPs, from close to $18,000 (including several different modalities of imaging-based testing, as of 1995)29 to near $3,500 to perform the Pathwork TOO Test.
One of the aims of this clinical verification study was to assess a known limitation of this assay: the possibility of an unknown primary cancer originating from a tissue site not covered by the panel of 15 tumor types used in the design of the Pathwork TOO Test. Theoretically, one might expect to obtain an indeterminate result when the actual tissue type does not belong to those covered by the Pathwork TOO Test. Our results showed that to be the case in 33% of the off-panel samples analyzed (2/6). However, when testing specimens that have been histologically classified as HNSCC (a tissue type that is not covered by the assay) the Pathwork TOO Test issued lung or bladder results in 71% of the cases (5/7). This finding is particularly interesting because although bladder (urothelial) carcinoma is not technically squamous, it is well known to express an immunohistochemical profile similar to that of squamous carcinomas, including such markers as CK5/6 and nuclear positivity for p63.
It should come as no surprise that there would be some overlap in the gene expression of squamous and urothelial carcinomas that would be picked up by the Pathwork TOO Test. Understanding this performance characteristic of the assay may be helpful when the differential diagnosis for a difficult case includes HNSCC and other tissue types covered by the 15-tissue panel, other than lung and bladder. Thus, one of our cases benefited from this test when a suspected thyroid origin was excluded (SS for thyroid <5). These results emphasize the importance of interpreting the results of this test, as well as most molecular diagnostic tests, in their clinicopathologic and radiologic contexts.
Even though our laboratory requires that biopsy specimens chosen for molecular testing include a high tumor representation, inevitably one may encounter unique samples harboring less than 60% tumor content that may be of value. In the series of 43 samples described herein, only 3 samples were considered inadequate owing to low tumor content (<60%), with one of them being a liver biopsy sample. The Pathwork TOO Test detected a liver gene expression profile, which was consistent with the biopsy site, rather than the actual tumor type, which was confirmed histopathologically to be a lymphoma. Because the other 2 inadequate samples had been diagnosed as CUP, a direct comparison between the Pathwork TOO Test and the original diagnosis could not be performed. This is one of the major limitations for any clinical assay intended to determine the TOO in CUP specimens.30 However, we found that the Pathwork TOO Test was able to detect the primary tissue type in 1 of the 5 adequate CUP samples analyzed in this study, while identifying a squamous cell type in 3 of them. Last, one of the CUP cases corresponding to a liver mass showed an immunophenotype that does not exclude a possible cholangiocarcinoma origin (positive staining for epithelial membrane antigen, Cam 5.2, and MOC-31),31 which could be misclassified as liver by the Pathwork TOO Test by virtue of cholangiocarcinoma not being a part of the 15-tissue panel covered by the assay or by the possibility of having a hepatocellular and cholangiocarcinoma mixed tumor. Our results for the off-panel and CUP samples clearly indicate that the tissue type and the cell type may be confounded in the results for the Pathwork TOO Test and that careful clinicopathologic assessment has to be made when interpreting such results.
Altogether, the results presented herein confirm the previously established clinical performance of this FDA-cleared, microarray-based gene expression assay and underscore the clinical usefulness and limitations of the test. The understanding of such limitations may assist pathologists during the interpretation of the assay’s results. This study verified the performance of the Pathwork TOO Test, which shows good potential as a valuable ancillary tool for pathologists for prompt and accurate diagnosis of poorly differentiated and undifferentiated tumor samples.
Supported by a sponsored research agreement with Pathwork Diagnostics, Sunnyvale, CA. Human tissues, patient consents, and clinical data were obtained through a collaboration with the Tissue and Data Acquisition and Analysis Core, supported by Cancer Center Support Grant No. NCI P30 CA016059 at the Massey Cancer Center, Richmond, VA, and Department of Pathology, Virginia Commonwealth University, in support of the Tissue Acquisition System to Support Cancer Research protocol.
References
Author notes
Drs Dumur and Fuller contributed equally to this work.